Introduction
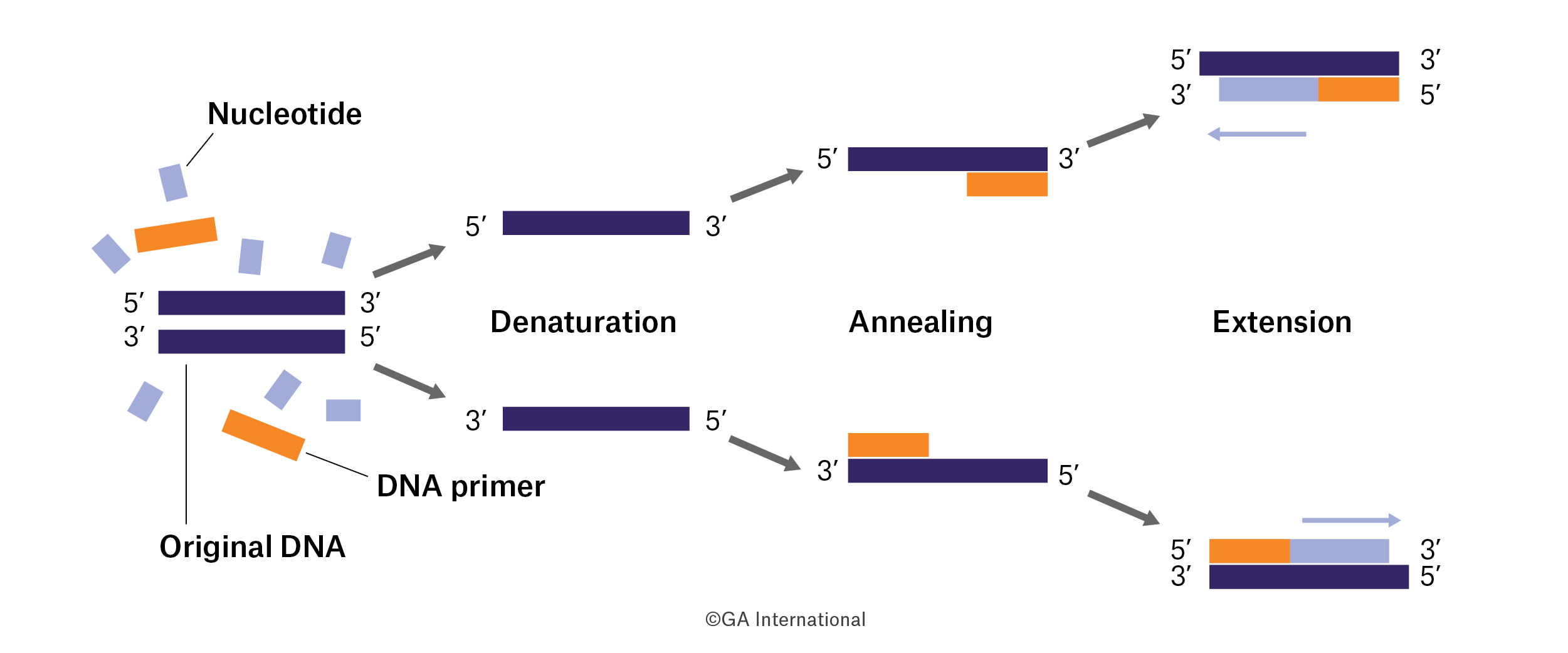
PCR (polymerase chain reaction) is a revolutionary method developed by Kary Mullis in the 1980s. Gentaur PCR is based on the use of DNA polymerase’s ability to synthesize a new DNA strand complementary to the offered template strand. Because DNA polymerase can add a nucleotide only to a preexisting 3′-OH group, it needs a primer to which it can add the first nucleotide. This requirement makes it possible to delineate a specific region of template sequence that the investigator wishes to amplify. At the end of the PCR reaction, the specific sequence will accumulate into billions of copies (amplicons).
PCR components
DNA Template: The DNA sample that contains the target sequence. At the beginning of the reaction, high temperature is applied to the original double-stranded DNA molecule to separate the strands from each other. DNA polymerase: a type of enzyme that synthesizes new DNA strands complementary to the target sequence. The first and most commonly used of these enzymes is Taq DNA polymerase (from Thermus aquaticus), while PfuDNA polymerase (from Pyrococcus furiosus) is widely used due to its higher fidelity in copying DNA.
Although these enzymes are subtly different, they both have two capabilities that make them suitable for PCR: 1) they can generate new DNA strands using a DNA template and primers, and 2) they are resistant to heat. DNA strands that are complementary to the target sequence. The polymerase starts to synthesize new DNA from the end of the primer. Nucleotides (dNTPs or deoxynucleotide triphosphates): individual units of bases A, T, G and C, which are essentially “building blocks” for new DNA strands. RT-PCR (reverse transcription PCR) is a PCR preceded by the conversion of the RNA sample into cDNA with the enzyme reverse transcriptase.
Limitations of PCR and RT-PCR
The PCR reaction begins to generate copies of the target sequence exponentially. Only during the exponential phase of the PCR reaction is it possible to extrapolate backwards to determine the initial amount of the target sequence contained in the sample. Due to polymerase reaction inhibitors in the sample, reagent limitation, accumulation of pyrophosphate molecules, and self-relation of the accumulated product, the PCR reaction ultimately fails to amplify the target sequence at a rapid rate. exponential and a “plateau effect” occurs. making endpoint quantification of PCR products unreliable. It is this attribute of PCR that makes quantitative real-time RT-PCR so necessary.